“The integration of artificial intelligence (AI), biomarkers, ageing biology, and longevity medicine stands as a cornerstone for extending human healthy lifespan.”
Imagine a future where we not only live longer but stay healthy throughout those extra years. Thanks to recent breakthroughs in biotechnology and artificial intelligence (AI) in healthcare, this vision is closer to becoming a reality.
Advancements in Aging Research
Aging research has made significant progress in recent years by combining disciplines like biology, technology, and medicine to tackle the challenges of extending healthspans and reducing age-related diseases. While people today live longer than ever before, extending our “healthspan”—the years we stay active and illness-free—remains challenging. AI and health biomarkers (biological indicators of our body’s condition) are now key tools in the pursuit of longer, healthier lives.
In a recent paper, led by corresponding authors Yu-Xuan Lyu from Southern University of Science and Technology Shenzhen; Alex Zhavoronkov from Insilico Medicine AI Limited, Masdar City, Abu Dhabi; Morten Scheibye-Knudsen and Daniela Bakula from the Center for Healthy Aging, University of Copenhagen, along with numerous other collaborators, the transformative potential of AI in aging research was explored. The research paper, titled “Longevity biotechnology: bridging AI, biomarkers, geroscience and clinical applications for healthy longevity,” was published as the cover paper in Aging’s Volume 16, Issue 20.
The Study: A New AI-Powered Approach to Aging
The work summarizes insights from the 2023 Aging Research and Drug Discovery Meeting. Researchers from renowned institutions explored how AI, biomarkers, and clinical applications can work together to enhance longevity. This fusion, termed “longevity biotechnology,” promises to transform healthcare from reactive treatments to proactive, preventive measures focused on staying healthy as we age.
The Challenge: Targeting Multiple Health Conditions with Longevity Biotechnology
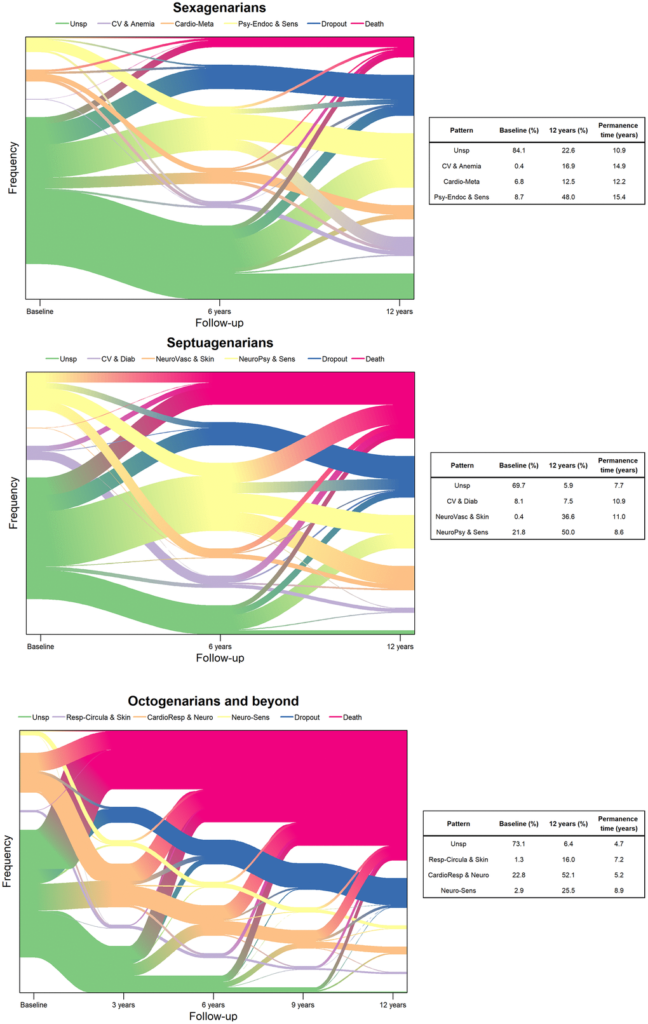
Traditional aging research often targets single diseases, but most elderly individuals experience multiple chronic conditions. Addressing this complex challenge requires identifying biological markers that indicate aging and predicting health risks before diseases manifest.
The Breakthrough: AI in Biomarker Discovery for Aging
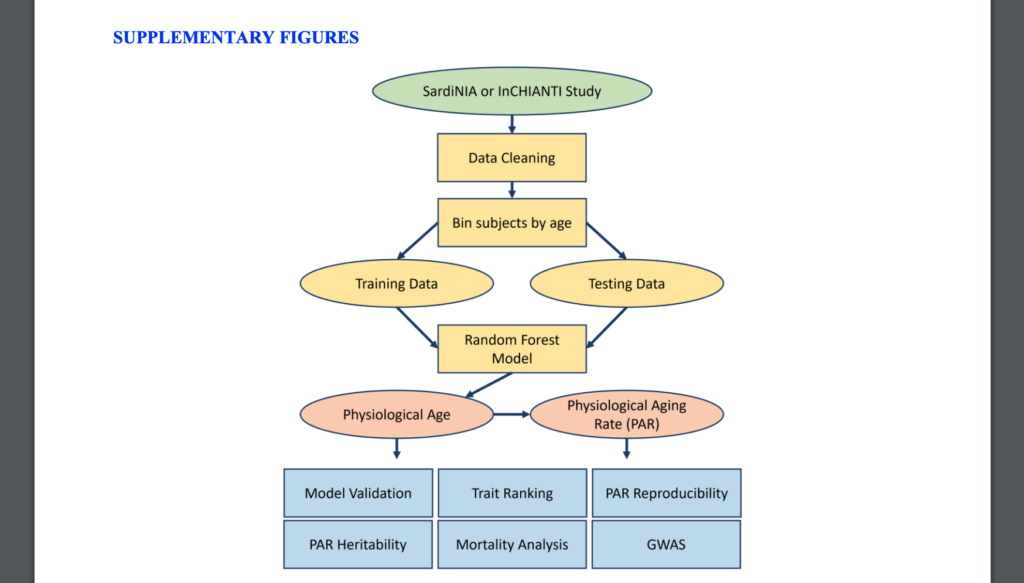
The study highlights how AI can accelerate the discovery of biomarkers, allowing scientists to understand aging at the cellular level. By using machine learning to identify unique patterns, researchers can estimate biological age, discover potential treatments, and evaluate the impact of lifestyle changes on health. This personalized approach enables healthcare providers to create prevention and treatment plans suited to each person’s unique health needs.
The Future of Healthcare: Preventive, AI-Driven Longevity Treatments
Currently, healthcare often focuses on managing diseases as they arise. However, these AI-driven tools could bring about a shift to preventive healthcare. Instead of waiting for age-related illnesses, clinicians could use AI insights to address aging’s root causes, improving health before issues arise.
While the promise of AI in healthcare is significant, the research team emphasizes that further investment is needed to make these AI-driven approaches accessible and accurate. With continued advancements, longevity biotechnology could become a standard part of healthcare, offering a new way to maintain vitality and well-being as we age.
Conclusion
Longevity biotechnology represents a groundbreaking shift, with AI and biomarkers helping us envision a future of healthier, longer lives. This approach brings us closer to understanding and managing the aging process, making extended healthspans a real possibility.
Click here to read the full research paper in Aging.
—
Aging is indexed by PubMed/Medline (abbreviated as “Aging (Albany NY)”), PubMed Central, Web of Science: Science Citation Index Expanded (abbreviated as “Aging‐US” and listed in the Cell Biology and Geriatrics & Gerontology categories), Scopus (abbreviated as “Aging” and listed in the Cell Biology and Aging categories), Biological Abstracts, BIOSIS Previews, EMBASE, META (Chan Zuckerberg Initiative) (2018-2022), and Dimensions (Digital Science).
Click here to subscribe to Aging publication updates.
For media inquiries, please contact [email protected].