“Aging is a multifaceted process influenced by intrinsic and extrinsic factors, with lipid alterations playing a critical role in brain aging and neurological disorders.”
A new study published recently as the cover of Aging Volume 17, Issue 6, describes a new method to estimate how fast the brain is aging. By analyzing lipids, or fat molecules, in brain tissue, researchers from the National University of Singapore and Hanze University of Applied Sciences created a biological “clock” called DoliClock. This innovation highlights how conditions such as autism, schizophrenia, and Down syndrome are associated with accelerated brain aging.
Understanding Brain Aging
As people grow older, their brains naturally change. However, in many neurological disorders, these changes seem to appear earlier and progress more rapidly. Disorders like autism, schizophrenia, and Down syndrome reduce quality of life and contribute to premature death. Scientists have long searched for better ways to measure biological age in the brain to understand these processes and develop strategies to slow them down.
Most existing methods for estimating biological age rely on genetic markers, such as DNA methylation, which are chemical modifications of DNA. While useful, these approaches may not fully capture the complexity of aging, especially in the brain. Lipids, which are essential components of brain cells and play important roles in energy storage and signaling, offer another perspective.
The Study: Building a Lipid-Based Aging Clock
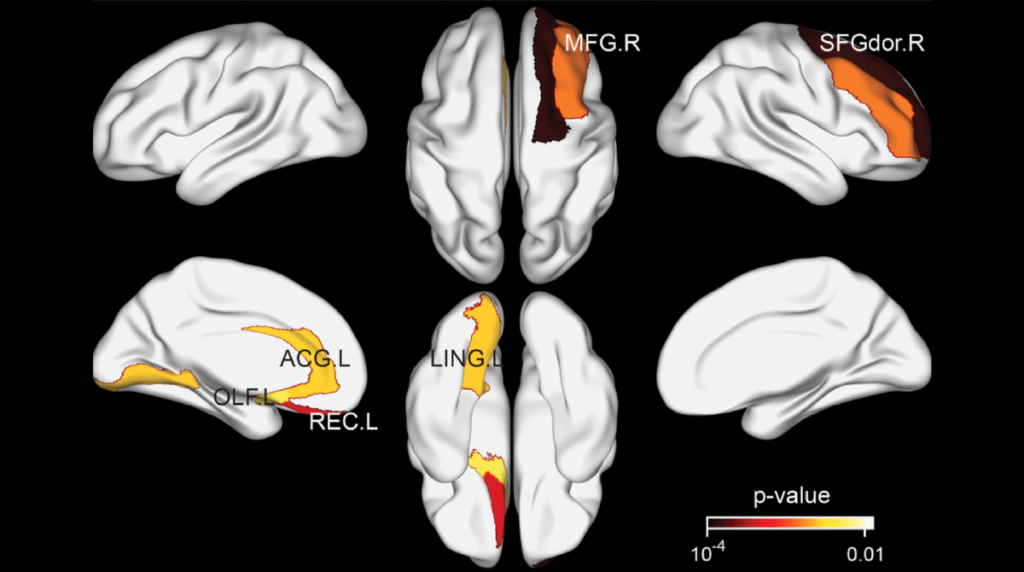
A team led by first author Djakim Latumalea and corresponding author Brian K. Kennedy introduced DoliClock, a model that predicts brain age using lipid profiles from the prefrontal cortex. This region of the brain, located just behind the forehead, plays a key role in decision-making, memory, and emotional regulation.
The study titled “DoliClock: a lipid-based aging clock reveals accelerated aging in neurological disorders” analyzed post-mortem brain samples from individuals with and without neurological conditions such as autism, schizophrenia, and Down syndrome.
The researchers focused on a class of lipids called dolichols, which are involved in vital cellular processes such as protein transport and glycosylation. These lipids tend to accumulate in brain tissue as people age, making them promising markers for measuring biological aging.
Results: Lipids Reflect the Pace of Aging
The DoliClock model showed that dolichol levels in the brain increased gradually with age. This change became particularly noticeable around the age of 40, suggesting a shift in how the brain regulates lipid metabolism during midlife. In addition to dolichols, the researchers observed an increase in entropy, a measure of disorder in lipid composition, which also intensified around this age.
When applied to brain samples from individuals with neurological disorders, DoliClock revealed significant differences. Samples from people with autism, schizophrenia, and Down syndrome showed higher predicted biological ages compared to their actual ages. This finding indicates that these disorders are associated with accelerated brain aging. The results align with previous studies using other biological clocks but add a new layer of understanding by focusing on lipid metabolism.
The Impact: A New Window into Brain Aging
DoliClock represents an important step in aging research because it demonstrates how lipid profiles can serve as markers of biological age. Unlike genetic markers, which may not fully capture brain-specific changes, lipidomic data directly reflect the brain’s structure and metabolic state. Dolichols, in particular, emerged as strong indicators of aging and may also play a role in the development of neurological disorders. This lipid-based clock could help scientists better understand the brain aging process and identify individuals at risk of premature decline.
Future Perspectives and Conclusion
DoliClock opens new possibilities for studying the molecular basis of brain aging. Although the current study used post-mortem brain tissue, future research could adapt this approach for use with more accessible samples. Similar lipid signatures might eventually be detectable in blood or cerebrospinal fluid, offering a non-invasive way to monitor brain health. Such tools could support early diagnosis and help track the effectiveness of treatments designed to slow brain aging.
Investigating how interventions such as dietary changes or medications affect lipid-based aging markers could also lead to new strategies for promoting healthy brain aging, making DoliClock a promising foundation for further exploration in aging research and brain health.
Click here to read the full research paper published in Aging.
___
Aging is indexed by PubMed/Medline (abbreviated as “Aging (Albany NY)”), PubMed Central, Web of Science: Science Citation Index Expanded (abbreviated as “Aging‐US” and listed in the Cell Biology and Geriatrics & Gerontology categories), Scopus (abbreviated as “Aging” and listed in the Cell Biology and Aging categories), Biological Abstracts, BIOSIS Previews, EMBASE, META (Chan Zuckerberg Initiative) (2018-2022), and Dimensions (Digital Science).
Click here to subscribe to Aging publication updates.
For media inquiries, please contact [email protected].